University of Toronto
University of Toronto
University of Alberta
University of Toronto
University of Toronto
IOCB Prague
IOCB Prague
🌟 Enveda Seedling Prize Winner and 2nd place overall at the Evolved Bio x ML Hackathon 2024 🌟

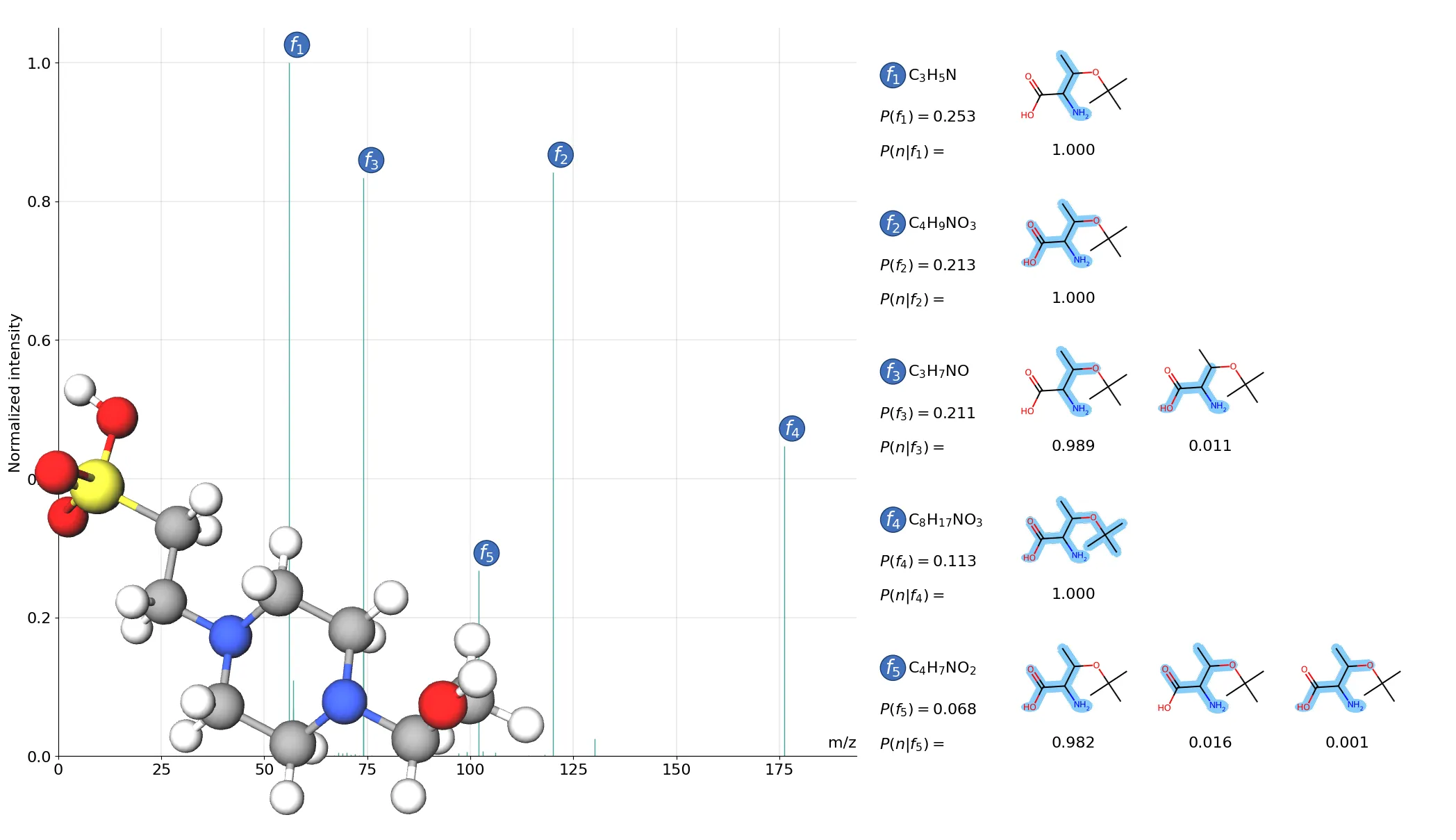
To accelerate scientific discovery, we need to identify a large number of compounds with high precision. MS/MS can do that. Going from spectrum to molecule is hard, 90% of the molecule spectra are unknown.
We think that requires three pillars: high-resolution prediction, interpretability, and incorporating large-scale unlabelled spectra data. In the current version, FraGNNet accurately predicts high-resolution spectra. FraGNNet also offers interpretability through annoting peaks with their possible fragmentation paths. In the next step we want to incorporate DreaMS, which contains information from 1000x times more data from unlabeled spectra.
We are working on a fleshed-out version of MSEffect that includes DreaMS, together with a full demo. Stay tuned!
@misc{young2024fragnnetdeepprobabilisticmodel,
title={FraGNNet: A Deep Probabilistic Model for Mass Spectrum Prediction},
author={Adamo Young and Fei Wang and David Wishart and Bo Wang and Hannes Röst and Russ Greiner},
year={2024},
eprint={2404.02360},
archivePrefix={arXiv},
primaryClass={cs.LG},
url={https://arxiv.org/abs/2404.02360},
}
@article{bushuiev2024emergence,
author = {Bushuiev, Roman and Bushuiev, Anton and Samusevich, Raman and Brungs, Corinna and Sivic, Josef and Pluskal, Tomáš},
title = {Emergence of molecular structures from repository-scale self-supervised learning on tandem mass spectra},
journal = {ChemRxiv},
doi = {doi:10.26434/chemrxiv-2023-kss3r-v2},
year = {2024}
}